
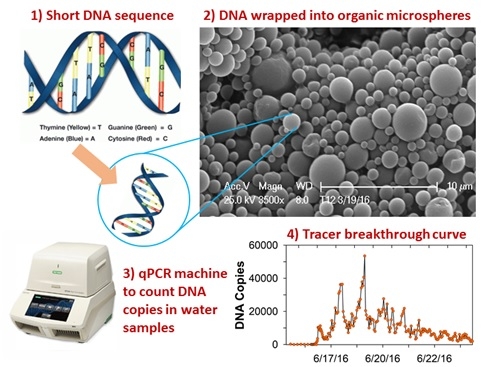
To overcome the problems that have plagued traditional tracer studies we have developed a new tracer concept that utilizes bio-molecular nanotechnology. We use short, artificially made DNA sequences that are wrapped into a safe, biodegradable polymer composed of polylactic acid (PLA) to protect the DNA from being eaten by microbes during the transport process. Because DNA is made of the four basic building blocks adenine (A), thymine (T), guanine (G) and cytosine (C) (Fig. 1), which can be combined in any random order, our unique DNA-based tracers allow, in theory, fabrication of an enormous number of unique tracers (approximately 1.61 x 1060) with identical transport properties. Because of their unique DNA sequence the tracers have unique IDs, thus, we can use multiple tracers at the same time in the same watershed. The amount of tracer present in a streamwater sample can be estimated with real-time quantitative polymerase chain reaction (qPCR), a method commonly used in molecular biology, which determines the number of DNA copies present in a sample. Finally, during the tracer fabrication process, we can alter the size of the tracer particles to anything from 200 nanometers (size of a virus) to 1 micrometer (size of colloids or bacteria), which helps mimicking the physical transport properties of the pollutant of interest.
In the next two years we will test differently sized DNA tracers in a well-studied, small experimental watershed at SFREC to test their use for identifying hydrologic flow pathways. This study will provide invaluable information for the understanding of processes in hydrologic systems that can be used to improve hydrological and biogeochmical models used to predict transport of pollutants from hillslopes to streams. To achieve this broader goal, we will distribute 5 different DNA tracers at 5 different locations at incrementally increasing distances from a trenched hillslope at SFREC and measure the tracer breakthrough curves in a runoff collection system and the watershed outlet. The injection locations will be chosen to represent a range of specific soil-landscape characteristics such as places with relatively deep soils, shallow soils and areas where flow visually concentrates in the landscape. With the tracer experiments we also hope to quantify preferential flow pathways (e.g. macropore flow) in the watershed. Preferential flow is hypothesized to lead to shorter travel times of water and pollutants through soils and the vadose zone. Because of the variable size of our DNA tracers (0.1-0.4 µm) they could be particularly useful for quantifying macropore and preferential flow because we expect that a small fraction of the tracers will be filtered out in the soil matrix while the majority of the DNA tracers will move along the most rapid flow pathways. The results from these experiments, if successful, will provide improved estimates of the time it takes for water and solutes to travel through the soil to streams, which will allow us to more accurately predict the risk of pollution of streams and surface water bodies during storm events.
